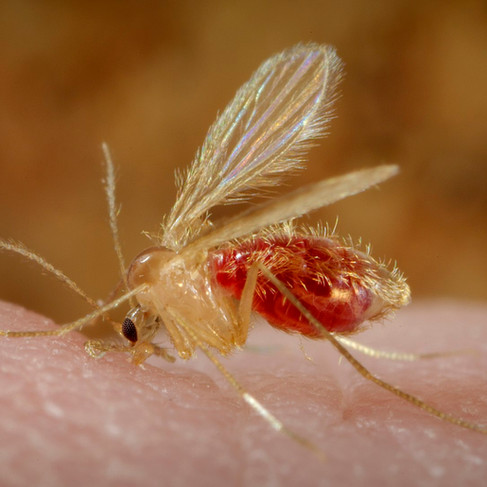
Sand flies: very small disease vectors
- Stephen Richards & Mary Ann McDowell
- Sep 20, 2022
- 3 min read
Blood feeding insects that feed on multiple people can spread disease. Mosquitoes are one example where the female mosquito requires a blood meal for protein to properly grow and lay her eggs. This act of blood feeding spreads the malaria parasites to and between people. Another example are sand flies. Sand flies, about ¼ the size of a mosquito, are tiny insect vectors of a different parasite – Leishmania.
Leishmaniasis is the second biggest parasitic killer after malaria. It is estimated to kill around 40,000 people every year, and accounts for about 2.4 million disability-adjusted life-years (DALYs). The Leishmania parasite is transmitted to humans when female sand flies bite humans to obtain blood meals that they require for reproduction. As of August 2022 (the time of writing), there is an outbreak of cutaneous leishmaniasis in Syria near a polluted and drying up river – as reported by VOA news here.
Current treatments of leishmaniasis are highly toxic and/or expensive and no efficacious vaccine exists. Therefore control of this disease requires controlling the Phlebotomine sand flies that transmit Leishmania parasites. A better understanding of sand fly biology and populations is necessary for the control and monitoring of the disease.
Today, we share two chromosome length assemblies for these two sandfly species: Phlebotomus papatasi and Lutzomyia longipalpis. P. papatasi is a vector of the cutaneous leishmaniasis causing parasite Leishmania major in the Old World. L. longipalpis is a vector transmitting Leishmania infantum that causes visceral leishmaniasis in South America.
Left: Lutzomyia longipalpis sandfly by Ray Wilson, (2009) PLoS Pathogens Issue Image - Vol. 5(8) August 2009. PLoS Pathog 5(8): ev05.i08. doi:10.1371/image.ppat.v05.i08, [CC BY 4.0]. Right: Phlebotomus papatasi sandfly by Frank Collins, Centers for Disease Control and Prevention's Public Health Image Library (PHIL), identification number #10277, [public domain].
It is our hope that these new high quality reference genomes will enable new and improve ongoing methods of integrated vector control. The improved genome assemblies should enable complex genetic analyses, including descriptions of species complexes, estimation of effective population sizes, monitoring of pesticide resistance allele emergence, and hopefully eventually lead to new leishmaniasis-control methods. Recently, new mosquito genomic resources enabled new vector control methods such as CRISPR-Cas9-based manipulation that create gene drives to prevent malaria transmission and microbiome manipulation with Wolbachia to prevent transmission of dengue. We hope that new reference genomes will enable similar innovative approaches to be developed for sand flies.
The extremely small size of the sand fly made generating these genomes a challenge. Only about 30-50 nanograms of DNA can be isolated from a single male sandfly (males are used here to characterize both the X and the Y sex chromosomes). Using pooled samples and older sequencing technologies in the early sequencing attempts produced extremely fragmented assemblies. We would like to thank researchers at Pacific Biosciences for using these species as a test for their new ultra-low input library generation protocol for the HiFi sequencing technology. When combined with the Hi-C magic performed by the DNA Zoo, we now have highly accurate contiguous sequences scaffolded to chromosome lengths that should stand as a reference for future leishmaniasis prevention research for years to come.
Data from two PacBio Sequel II SMRT cells generated at the BYU sequencing center was assembled with hifiasm (Cheng et al., 2021). Pacific Biosciences generated HiFi data for Phlebotomus papatasi, the draft assembly for was generated by Dr. Sarah Kingan at Pacific Biosciences. For Lutzomyia longipalpis, Sequel II Hifi Data was generated by Oanh Nguyen at the UC Davis Genome Center from a single male using the ultra-low input DNA library kit generated by Dr Kingan and Colleagues at PacBio, and the initial assembly was performed by Stephen Richards using hifiiasm. These high-quality drafts were upgraded to chromosome-length assemblies using Hi-C data from multiple male individuals from the colonies at The University of Notre Dame maintained by Drs. Mary Ann McDowell and Douglas Shoue.
Check out the interactive Juicebox.js instance below for a contact map of five P. papatasi and four L. longipalpis chromosomes, and visit the assembly pages (here and here) for more information and details on the procedure. The genome assemblies are also available on NCBI as Ppap_2.0 (P. papatasi) and ASM2433408v1 (L. longipalpis).
Blog post by Stephen Richards and Mary Ann McDowell.